Co-registering EEG Data
Overview
Teaching: 20 min
Exercises: 10 minQuestions
Why is co-registering EEG data important?
How is co-registeration of EEG data done?
Objectives
Understand why co-registering data is important.
Understand how to co-register EEG data.
Introduction
Co-registering is the process of aligning the recording montage with a standard montage, allowing for the data to be compared to other studeis. Co-registration of the data to the standard MNI head will occur during the Lossless pipeline in the s01_scalpart script. Prior to running the pipeline, the appropriate transformation matrix that will warp the recorded data to the standard montage needs to be determined. The transformation matrix can be determined using the ll_validate.m script and this matrix can then be input into the [montage_info] field in the batch configuration file c01.
Co-registration Procedure
-
Open MATLAB and navigate to your local project directory to make it your current path.
-
Open EEGLAB by typing the following into the command window:
>> addpath derivatives/EEG-IP-L/code/install >> lossless_path >> eeglab -
To determine the correct transformation matrix to co-register your data to a standard montage (the MNI head) run the
ll_validate.mscript in the MATLAB command window:>> ll_validate -
In the open file dialog window navigate to
sub-001/eeg/and select thesub-001_task-FaceFO_eeg.edffile. You may need to change theFiles of Typefield toAll Files. Determining the transformation matrix only needs to be done once per study and this process can be completed using any of the data files. -
A popup window that plots the recording montage and the standard montage will appear. A mesh head will also be plotted with the montages.
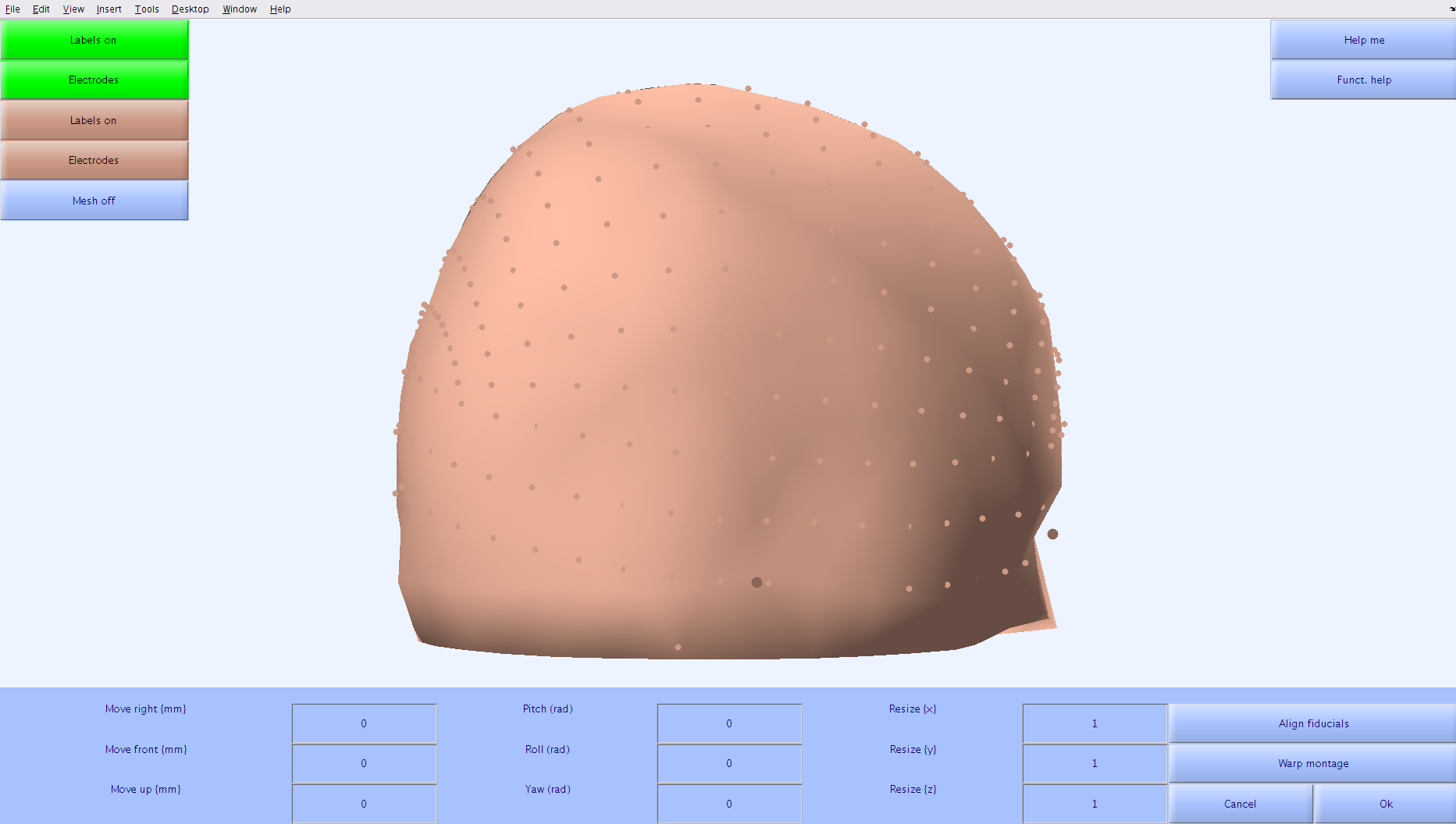
Determining the Transformation Matrix
The goal is to have the two montages sit on the same surface with corresponding electrodes as close to each other as possible. This can be done by adjusting the parameters of the transformation matrix that are located at the bottom of the window. The fiducials (LPA, Nz, RPA) should be used to determine if the montages are aligning. The electrode positions for the recording montage are shown in green and the electrode positions for the standard montage are shown in brown.
Hints
- The recording montage is on a very different scale than the standard montage. This can be adjusted by putting large numbers (>1000) in the
Resize(x),Resize(y), andResize(z)fields. - The nose direction of the recording montage is different than the nose direction of the standard montage. This can be corrected by putting (-1.58) in the
Yaw(rad)field. - Once the montages the same size and facing the same direction, use the
Move right,Move front, andMove upfields to adjust where the montages are in space relative to each other. - The mesh head that is plotted with the montages can be toggled on/off by pressing the
Mesh on/offbutton.
Solution
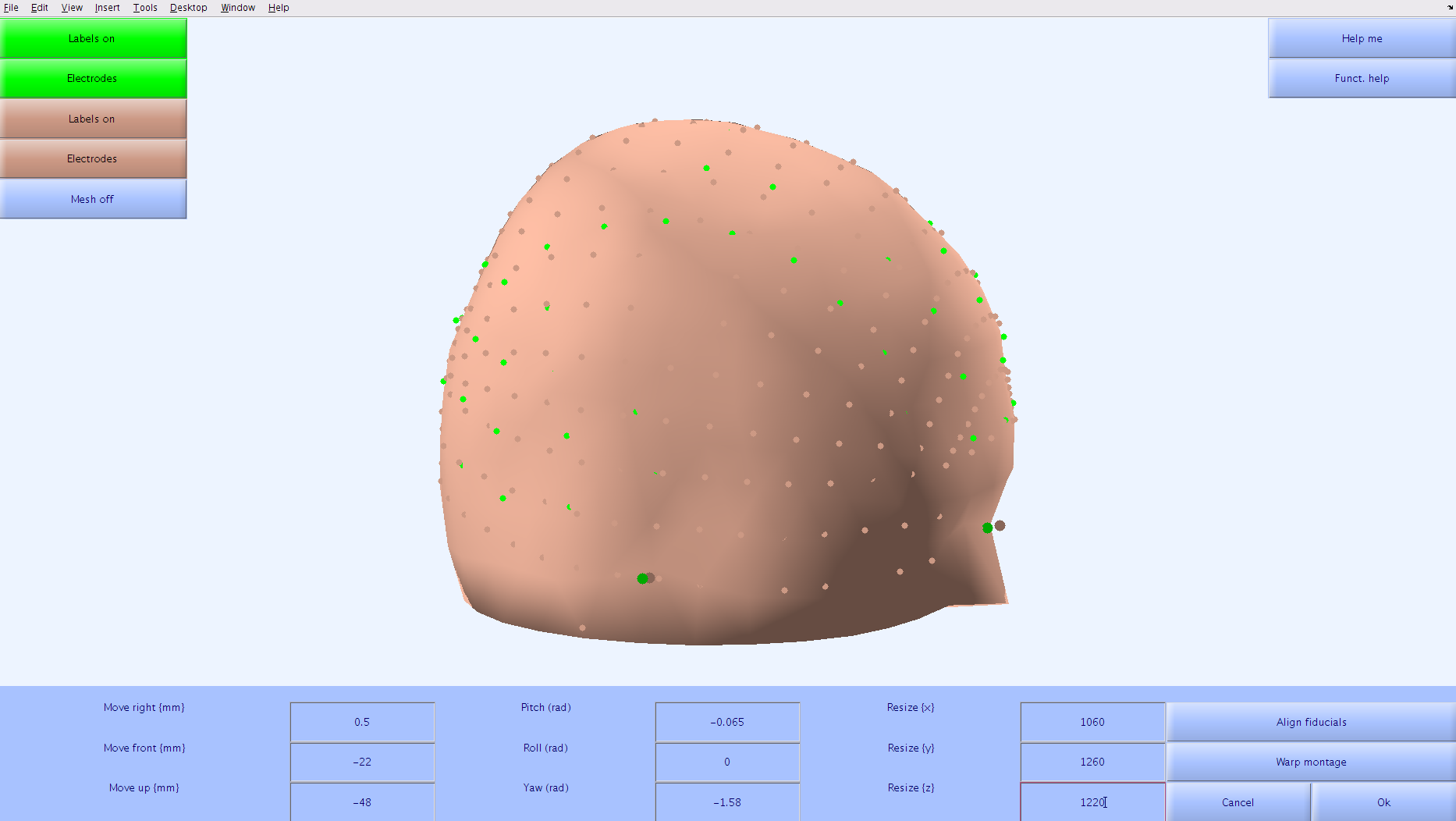 For the Face13 tutorial, the transformation matrix that will be input into
For the Face13 tutorial, the transformation matrix that will be input into [montage_info]inc01_scalpart.cfgis: [0.500 -22.000 -48.000 -0.065 0.000 -1.580 1060.000 1260.000 1220.000] - The recording montage is on a very different scale than the standard montage. This can be adjusted by putting large numbers (>1000) in the
-
When you are satisfied with the alignment between the two montages, select the
Okbutton. -
In the MATLAB command window, it will print: [montage_info] for c01_scalpart.cfg followed by the transformation matrix (nine numbers). Copy and paste these values into the
[montage_info]field in thec01_scalpart_remote.cfgfile located inderivatives/EEG-IP-L/code/config/face13_sbatch. These numbers need to be within square brackets in the batch configuration file.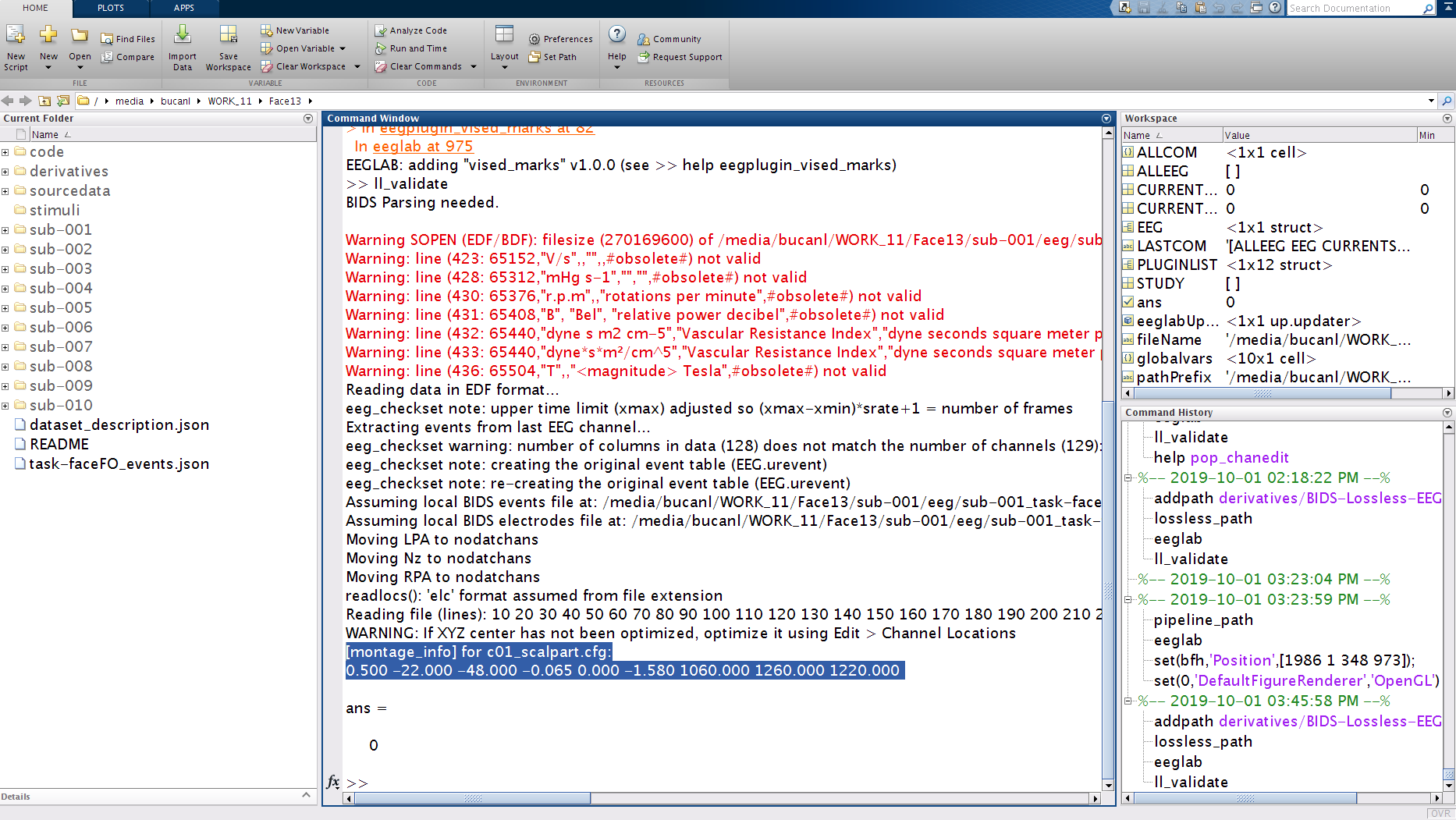
-
In the EEGLAB drop-down menu, navigate to File and select
Clear study/ Clear allto remove the dataset from memory.
Key Points
Co-registering data to a standard montage allows for comparison to other studies.
Co-registration occurs during the Lossless Pipeline in the s01 script based on a determined transformation matrix.