Running the Lossless Pipeline
Overview
Teaching: 35 min
Exercises: 10 minQuestions
How do we run a batch of EEG files through the Lossless pipeline?
How do we submit Lossless pipeline jobs to a remote parallel computing cluster?
Objectives
Submitting a batch of EEG files to run remotely through the Lossless pipeline.
Copy files from local to remote
-
In a terminal window opened to your local drives, navigate to your local project directory:
>> cd path/to/project/directory/Face13/ -
Copy the local subject folders to the remote computer cluster, leaving these files in BIDS format (
sub-*/eeg/). Again, replace [user_name] with your own username and [project_name] with your project name (the project name for the tutorial is ‘Face13’):>> rsync -rthvv --prune-empty-dirs --progress --include="*sub*" --include="*/" --exclude="*" --exclude="/*/*/*/*/" sub-* [user_name]@gra-dtn1.computecanada.ca:/scratch/[user_name]/[project_name]/Using gra-dtn1
For transfering files to and from the remote it is recommended to use the
gra-dtn1.computecanada.calogin node because it is dedicated to data transfer.Windows Support
Windows does not have support for rsync. Use the following command instead:
>> scp -r sub-* [user_name]@gra-dtn1.computecanada.ca:/scratch/[user_name]/[project_name]/Running Your Own Data- Staging Script
The staging script allows for additions to datafiles before they enter actual processing, such as the addition of in-task and break period annotations. If you are running your own data, a staging script can be customized for your dataset. The custom staging script would then need to be uploaded onto Graham where it will run during s01. More information on customizing a staging script can be found in the reference manual on the EEG-IP-L wiki. To upload your custom staging script to Graham, use the following command and update [staging_script] with the file name of your staging script as well as [user_name] and [project_name] with the appropriate names:
>> scp derivatives/EEG-IP-L/code/scripts/[staging_script] [user_name]@gra-dtn1.computecanada.ca:/scratch/[user_name]/[project_name]/derivatives/EEG-IP-L/code/scripts
Configuration file setup
-
Open MATLAB and navigate to your local project directory to make it your current path.
-
Open EEGLAB by typing the following into the command window:
>> addpath derivatives/EEG-IP-L/code/install >> lossless_path >> eeglab -
In the EEGLAB drop-down menu, navigate to File->Batch->Context Configuration and click
| Load context config |to load a default configuration file that can then be modified and saved. To load the deafult context configuration file, navigate to thederivatives/EEG-IP-L/code/config/directory and select the file namedcontextconfig.cfg.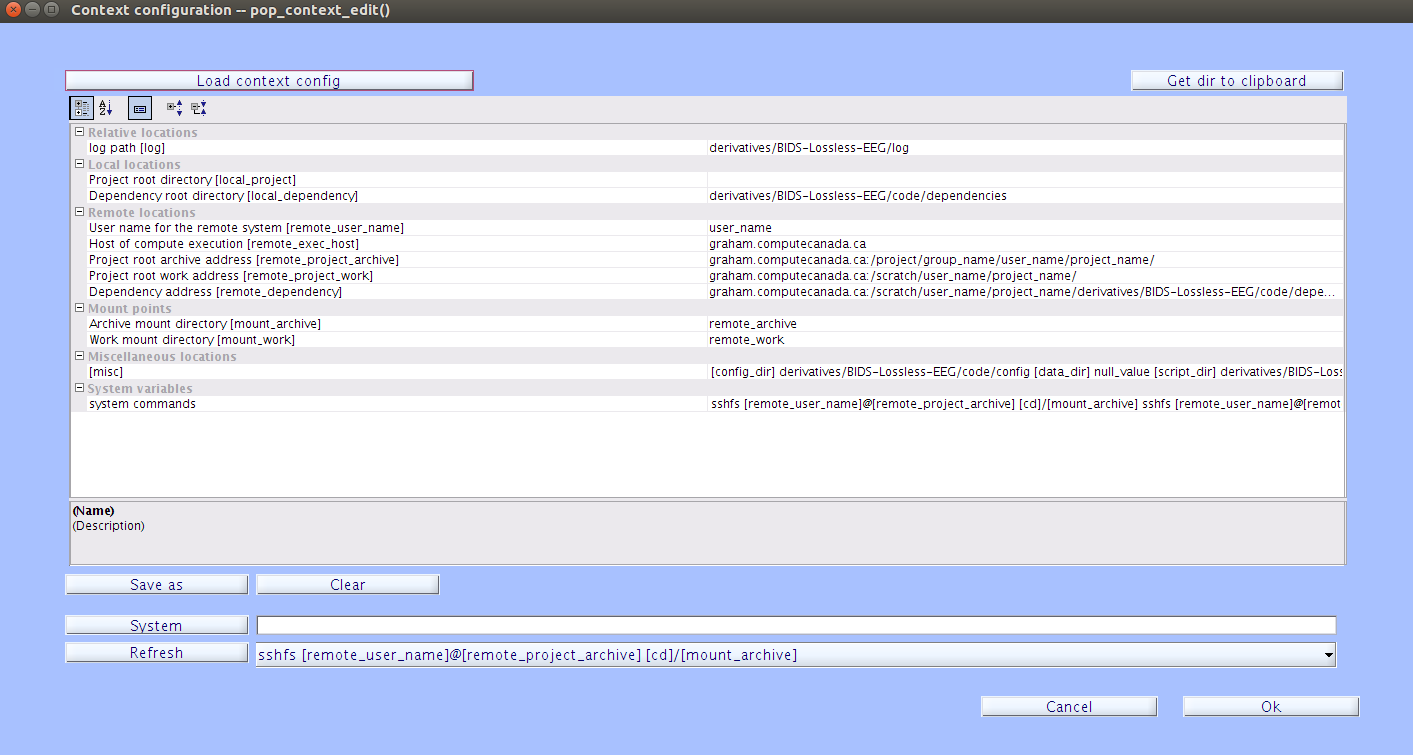
-
Modify the appropriate fields under Remote Locations, which will correspond to the remote paths for the project. Change the
remote_user_namefield to you user name on the remote system. The[remote_project_archive]field can be left as the default. In theremote_project_workandremote_dependencyfields, change the[user_name]and[project_name]to your remote user name and your project name. Once you have finished modifying the context configuration file, select| Save as |and navigate to thederivatives/EEG-IP-L/code/config/directory and save the configuration file here. It is recommended to save the context configuration file ascontextconfig_[user_name]_[project_name].cfg.Note
The
remote_exec_hostfield is the host and domain of the remote system. Theremote_project_archivefield is the remote path to the archieve location for where you would like to store processed files long term. On Graham, typically this is the project file system. Finally, theremote_project_workdirectory is the remote path to the location of the work root project directory, where the actual jobs will be run, and theremote dependencyis the same as theremote_project_workdirectory, but a few levels deeper (derivatives/EEG-IP-L/code/dependencies/). For more info, see the Batch Context wiki about Context configuration files. -
Once you have finished modifying the context configuration file, press enter or click off of the current field. Then select
| Save as |and navigate to thederivatives/EEG-IP-L/code/config/directory and save the configuration file here. It is recommended to save the context configuration file ascontextconfig_[user_name]_[project_name].cfg. -
In the EEGLAB drop-down menu, navigate to File->Batch->Batch Configuration and click
| Get batch config file names |to load the default batch configuration file(s) that can then be modified and saved. The configuration files we want to edit for the Face13 data are namedc01-c05and are located in thederivatives/EEG-IP-L/code/config/face13_sbatchfolder. It is recommended that you load, edit and save the configutation files one at a time. -
Once you have a file selected, click the
| Clear/Load |button to load the file into the property grid.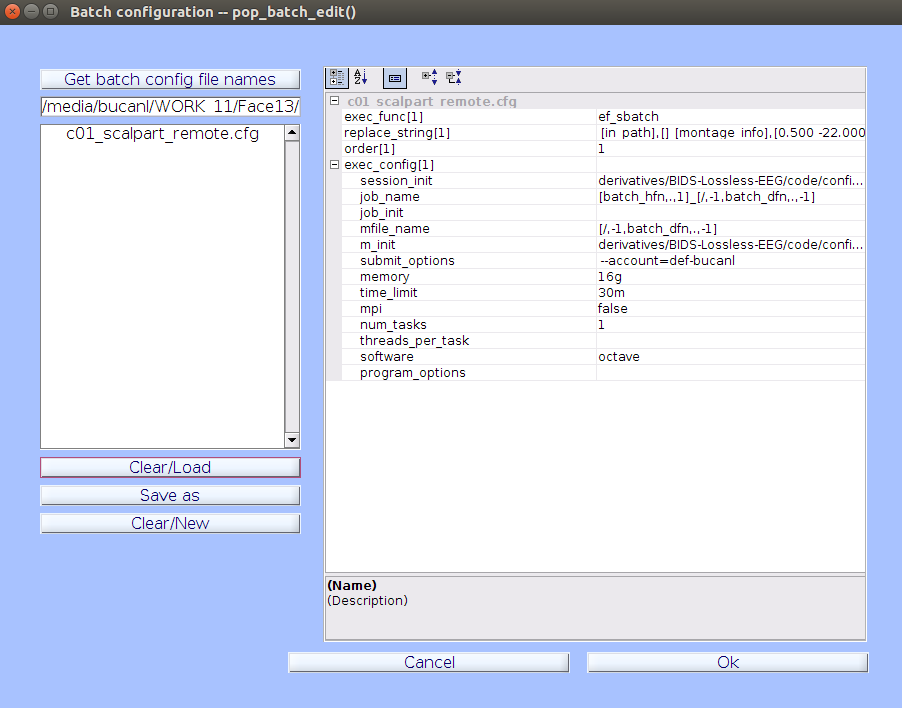
-
Change the
submit_optionsfield in each batch configuration file to--account=[group_name], where [group_name] is the name of your allocation group on Graham. The rest of the parameters in batch configuration files are optimized for the Face13 tutorial dataset.Running Your Own Data- Optimizing Parameters
Each script in the Lossless pipeline has parameters that relate to outlier detection criteria, montage information, and path specifications. Prior to running your own data through the Lossless pipeline, optiminal parameters should be determined. More information about the parameters in each script of the pipeline can be found on the EEG-IP-L wiki. The Lossless Pipeline Parameter Tutorial can be used to learn about the outlier dectection criteria and how to determine appropriate parameter values for your dataset.
-
Once you are done editing the parameters, press enter or click off of the current field. Then you can click
|Save as|and navigate back to thederivatives/EEG-IP-L/code/config/face13_sbatchfolder and save the configuration file with the same name as before.Note
If you have loaded and edited multiple configuration files at once, after you click
| Save as |, you must select all of the files and iteratively save each file. -
Click the
| Clear/New |button and repeat the process for all seven of the batch configuration files (c01-c05).
Common Errors
An easy way to check for errors is to compare your newly created files against the default Face13 configurations on the BUCANL Github. In particular, the
ordersubfield of each config file should remain exactly the same as the corresponding default file. If these have changed, you have most likely overwritten some of your config files. In this case, you will have to update these files and go through the editing process again. To update the files, you can copy and paste directly from the Github page to the configuration file in your project.
Submit jobs
-
In the EEGLAB drop-down menu, navigate to File->Batch->Run history template batch.
-
If your context configuration file is not already loaded, click
| Load context config |and load the context configuration file you saved in step 3 above. -
If your batch configuration files are not already loaded, click
| Load batch config |and then click| Batch configuration file |on the pop-up window. The configuration files we want to select for the Face13 data are in thederivatives/EEG-IP-L/code/config/face13_sbatchfolder. You want to select the seven files that are named ‘c01-c05’.Editing Config Files in a History Template Batch
If you have made any changes to the context config or batch config files, make sure you press enter or click off of the current field. This will ensure any changes you have made are saved.
-
Click
| History file |and load all the scripts (s01-s05) corresponding to each of the batch configuration files (c01-c05). These scripts are located in thederivatives/EEG-IP-L/code/scripts/directory. -
Open up a terminal window, and navigate to your local project directory again:
>> cd path/to/project/directory/Face13/ -
List all the data files you’d like to run through the pipeline. This can be done using the find command by typing:
>> find sub-* -type f -name "*_eeg.edf" -
This will print a list of all your initialized
*.edffiles, including the path, which you can then copy straight from the terminal into the file field in the run history template batch window, with one path/filename per line. -
Click
| Ok |and type your Graham password in the command window when prompted. You will have to enter your password several times, unless you have ssh keys set up. Your jobs should now start submitting for each data file, sequentially, one script at a time.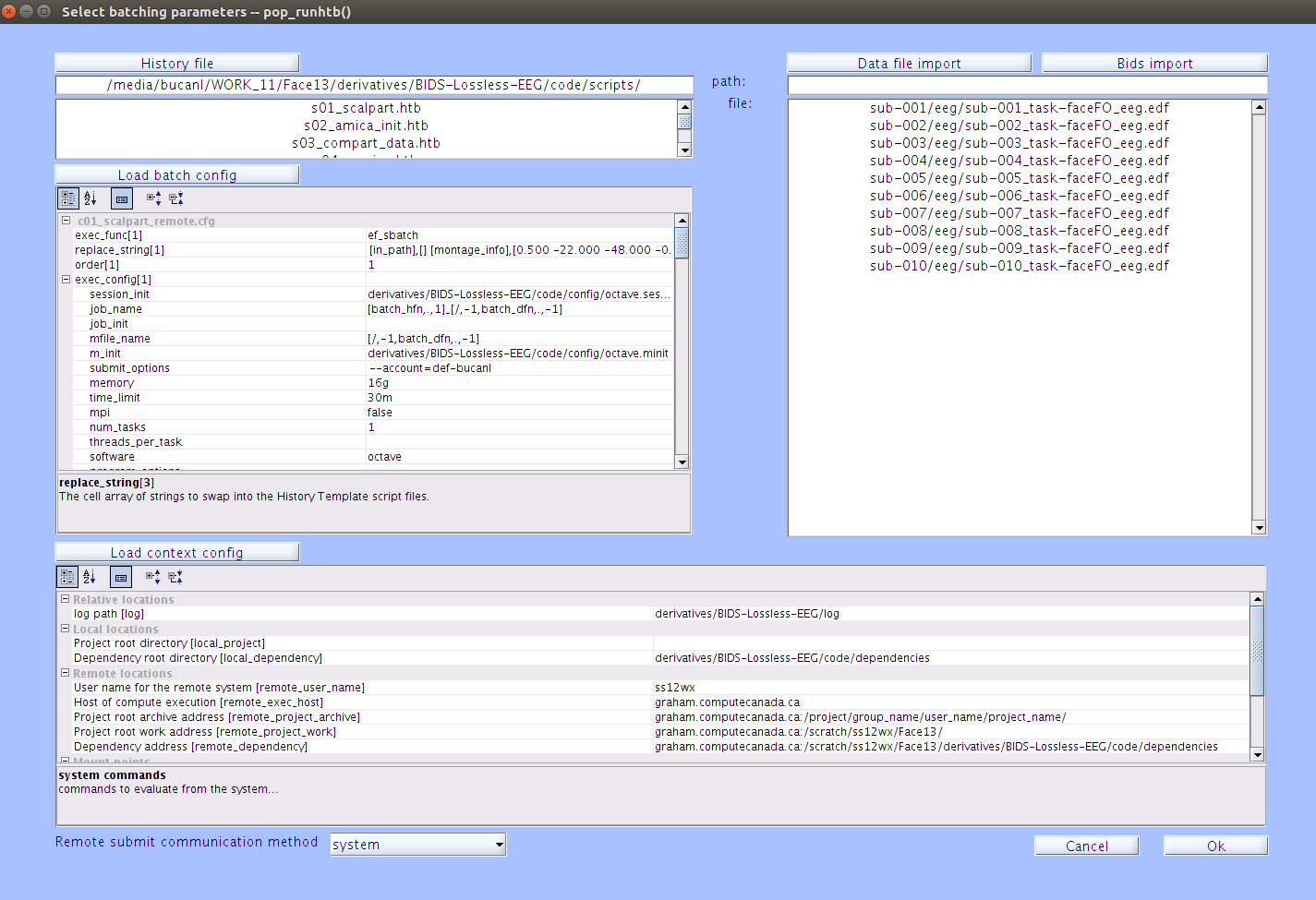
Query running jobs
-
To check the status of your submitted jobs, go to a terminal window and log into Graham again:
>> ssh [user_name]@graham.computecanada.ca -
To list all currently queued and running jobs and check their status, type:
>> squeue -u [user_name] -
To check if any particular file succeeded or failed running during a particular script, you may check the corresponding
*.logfile in thederivatives/EEG-IP-L/log/folder. A stack trace of any error will be printed here.
Copy files from remote to local
-
Once the files have successfully run through each stage of the pipeline, you should end up with an identical folder structure (
sub-*/eeg/) in your remotederivatives/EEG-IP-L/folder for each of the data files that ran through the pipeline. There will be intermediary files and several final output files for each participant. The final output files are*_ll.set,*.edf,*icasphere.tsv,*icaweights.tsv,*dipole.mat,*_annotations.mat,*_annotations.tsv,*_annotations.json, and*_iclabel.mat. -
To check if all files have in fact made it through the entire pipeline, you may locate these
*.edf*files using the find command, and seeing if there are any files missing:>> find derivatives/EEG-IP-L/sub-* -type f -name "*_ll.set*"Missing Files
Files that are missing from this find command did not make it all the way through the pipeline. The log files for the job submission can be investigated to determine why a file did not successfully complete. These log files are located in
derivatives/EEG-IP-L/log/. -
Now, you may copy these output files back to your local project directory. Make sure you are currently in your local project directory, if you aren’t already:
>> cd path/to/project/directory/Face13/ -
Now, transfer the files using the following command in the terminal, replacing [user_name] with your own username and [project_name] with ‘Face13’:
>> rsync -rthvv --prune-empty-dirs --progress --include="*dipole.mat" --include="*iclabel.mat" --include="*.edf" --include="*icaweights.*" --include="*icasphere.*" --include="*_annotations*" --include="*_ll*" --include="*/" --exclude="*" --exclude="/*/*/*/*/" [user_name]@gra-dtn1.computecanada.ca:/scratch/[user_name]/[project_name]/derivatives/EEG-IP-L/sub-* derivatives/EEG-IP-L/Windows Support
Windows does not have support for rsync. Use the below command instead. Note that more files will be downloaded as a result of scp grabbing all files in the subject folders.
>> scp -r [user_name]@gra-dtn1.computecanada.ca:/scratch/[user_name]/[project_name]/derivatives/EEG-IP-L/sub-* derivatives/EEG-IP-L/ -
Once this procedure is completed, you should notice all the output files in your local
derivatives/EEG-IP-L/directory. These files can now be put through the QC procedure for further processing.
Key Points
Ensure you have all the pipeline files and your data files on both the local and remote machine.
The input file for the Lossless pipeline is
*_eeg.edfand the output files are*.edf,*_ll.set,*icasphere.tsv,*icaweights.tsv,*dipole.mat,*_annotations.mat,*_annotations.tsv, and*_annotations.json.Remember to always pay attention to whether you are running a bash command on your local machine versus the remote computer cluster.